1.7 Working directories
The working directory is the default location where R will look for files you want to load and where it will put any files you save. One of the great things about using RStudio Projects is that when you open a project it will automatically set your working directory to the appropriate location. You can check the file path of your working directory by looking at bar at the top of the Console pane. Note: the ~ symbol above is shorthand for /Users/nhy163/ on a Mac computer (the same on Linux computers).
You can also use the getwd() function in the Console which returns the file path of the current working directory.
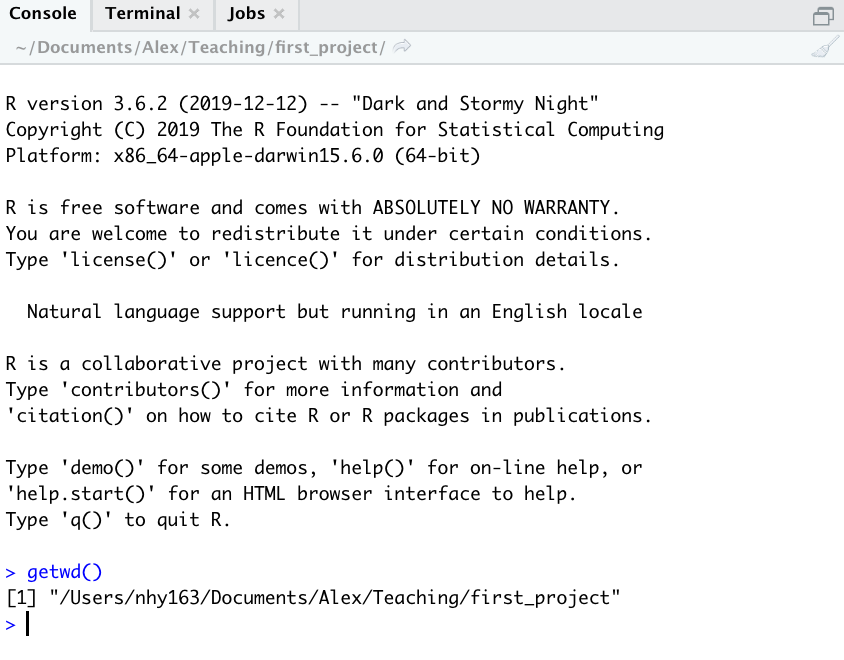
In the example above, the working directory is a folder called ‘first_project’ which is a subfolder of “Teaching’ in the ‘Alex’ folder which in turn is in a ‘Documents’ folder located in the ‘nhy163’ folder which itself is in the ‘Users’ folder. On a Windows based computer our working directory would also include a drive letter (i.e. C:/Users/nhy163/Documents/Alex/Teaching/first_project).
If you weren’t using an RStudio Project then you would have to set your working directory using the setwd() function at the start of every R script (something we did for many years).
However, the problem with setwd() is that it uses an absolute file path which is specific to the computer you are working on. If you want to send your script to someone else (or if you’re working on a different computer) this absolute file path is not going to work on your friend/colleagues computer as their directory configuration will be different (you are unlikely to have a directory structure /Users/nhy163/Documents/Alex/Teaching/ on your computer). This results in a project that is not self-contained and not easily portable. RStudio solves this problem by allowing you to use relative file paths which are relative to the Root project directory. The Root project directory is just the directory that contains the .Rproj file (first_project.Rproj in our case). If you want to share your analysis with someone else, all you need to do is copy the entire project directory and send to your to your collaborator. They would then just need to open the project file and any R scripts that contain references to relative file paths will just work. For example, let’s say that you’ve created a subdirectory called raw_data in your Root project directory that contains a tab delimited datafile called mydata.txt (we will cover directory structures below). To import this datafile in an RStudio project using the read.table() function (don’t worry about this now, we will cover this in much more detail in Chapter 3) all you need to include in your R script is
Because the file path raw_data/mydata.txt is relative to the project directory it doesn’t matter where you collaborator saves the project directory on their computer it will still work.
If you weren’t using an RStudio project then you would have to use either of the options below neither of which would work on a different computer.
setwd("/Users/nhy163/Documents/Alex/Teaching/first_project/")
dataf <- read.table("raw_data/mydata.txt", header = TRUE, sep = "\t")
# or
dataf <- read.table("/Users/nhy163/Documents/Alex/Teaching/first_project/raw_data/mydata.txt", header = TRUE, sep = "\t")For those of you who want to take the notion of relative file paths a step further, take a look at the here() function in the here package. The here() function allows you to automagically build file paths for any file relative to the project root directory that are also operating system agnostic (works on a Mac, Windows or Linux machine). For example, to import our mydata.txt file from the raw_data directory just use
library(here) # you may need to install the here package first
dataf <- read.table(here("raw_data", "mydata.txt"),
header = TRUE, sep = '\t',
stringsAsFactors = TRUE)
# or without loading the here package
dataf <- read.table(here::here("raw_data", "mydata.txt"),
header = TRUE, sep = '\t',
stringsAsFactors = TRUE)